Setting the Stage: From a Single Agent to a Team of Specialists
This is the third post in our ongoing series, Pharmacometrics × AI, where we're building an AI system to automate and augment complex pharmacometric workflows. Each post brings us closer to our final goal: an AI agent capable of performing these tasks securely and reliably. As we go I will be updating the PharmAI GitHub repository and would encourage anyone interested to jump in and start playing with the code.
To get you up to speed, here's where we've been so far:
- Part 1: The Vision: We established the "why" by identifying the core bottleneck in pharmacometric workflows—the manual, "iterative hell"—and proposed AI agents as the solution.
- Part 2: The Foundation: We provided the "how" by building our first "agentic atom," a single, capable agent demonstrating the core principles of security, reliability, and intelligence.
In our last post, we built a powerful single agent. However, that approach revealed a fundamental scaling limitation: a monolithic agent that handles all logic becomes increasingly complex, brittle, and difficult to optimize as requirements grow.
Today, we're tackling that head-on. We will architect a system with a main "Orchestrator Agent" that manages a team of specialized, independent "Tool Agents." By the end of this article, you will understand how to design a modular, composable system using a primary coding agent (Orchestrator), the Model Context Protocol (MCP), and DSPy (for tool develpment) to build and manage a suite of specialized pharmacometric AI tools.
The New Architecture: From Monolithic Agent to Composable System
The solution to our scaling problem lies in orchestration and composability. Rather than building one agentic workflow to do everything, we will architect a system of interchangeable components that can be combined to create complex workflows from simple, robust, and reusable parts.
This architectural shift mirrors proven patterns in software engineering, where microservices have replaced monolithic applications. This modularity is critical in regulated environments, as it allows individual components (e.g., a document parsing tool) to be optimized, updated, or replaced independently without requiring re-validation of the entire system. As noted in Anthropic's guide on Building Effective Agents, the key is designing systems that can evolve and scale without becoming unwieldy.
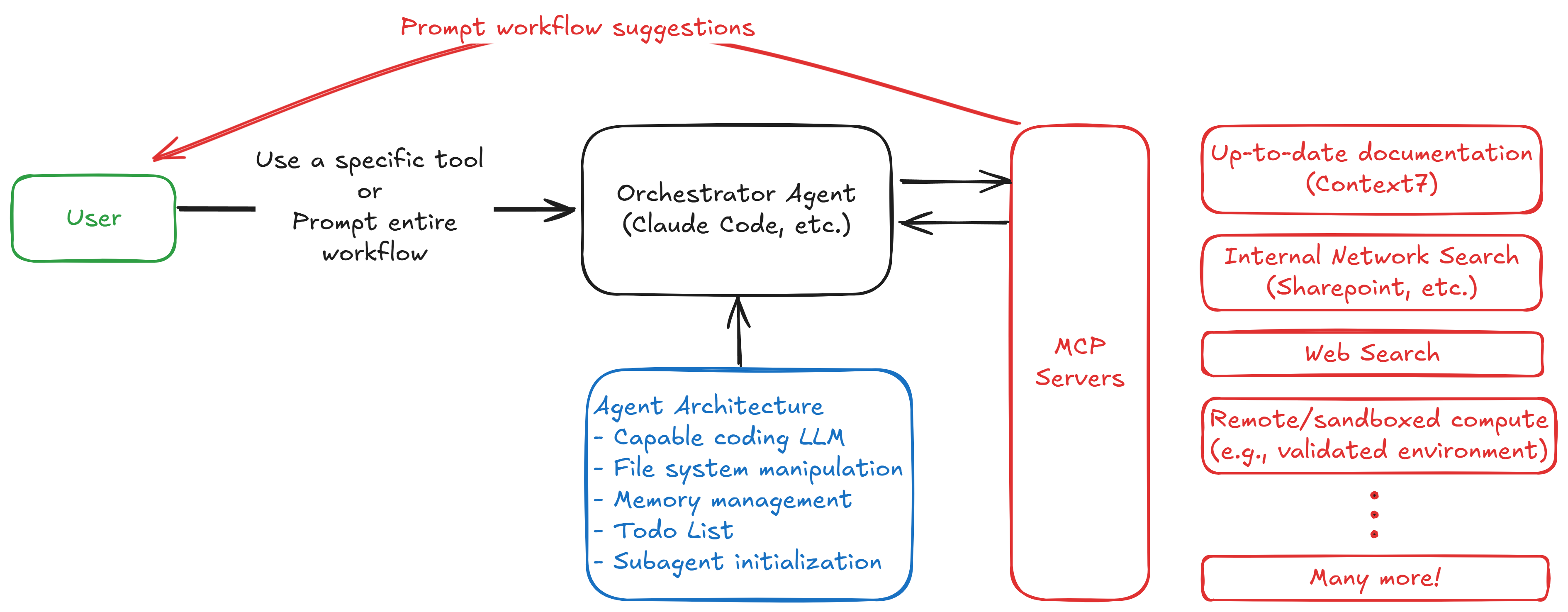
Here we have a core orchestrator agent that acts as the central hub, managing a team of specialized agents (tools) that perform specific tasks. This modular architecture allows us to:
- Scale: Add new tools or replace existing ones without disrupting the entire system.
- Evaluate: Continuously assess the performance of individual tools and the overall system, enabling data-driven improvements.
- Optimize: Focus on optimizing each tool independently, rather than trying to optimize a monolithic agent. Then we can optimize the orchestrator to effectively manage these tools using things like DSPy, fine-tuning, and reinforcement learning.
The Communication Backbone: Model Context Protocol (MCP)
The first challenge in our orchestrated system is establishing reliable communication between the orchestrator and our custom tools. We need a standard interface that ensures compatibility and maintainability.
Enter the Model Context Protocol (MCP), an open standard that acts as a universal adapter for AI tools. MCP allows any compliant agent (our orchestrator) to connect seamlessly with any compliant tool (our specialized agents). Think of it as USB-C for AI tools—a standardized connection that enables plug-and-play functionality.
The true power of MCP is that an MCP server can be a specialized agent itself, creating unprecedented composability. We can deploy MCP servers for:
- Task Parsing: An agent that reads pharmacometric analysis documents and generates structured plans (we'll build that below).
- Secure Code Execution: An agent that runs R code in controlled environments, from local REPLs to remote systems (inspired by tools like Code Sandbox MCP).
- Documentation Retrieval: Integration with community tools like Context7 to provide real-time, structured documentation for our tools.
- Compliance and Auditing: An agent that logs every tool call to create a comprehensive audit trail for regulatory submissions.
The Orchestrator: An Augmented LLM at the Center
Orchestrators were introduced in Anthropic's Building Effective Agents guide as a way to manage complex workflows by coordinating multiple agents and tools. The orchestrator acts as the "brain" of our system, receiving high-level goals and breaking them down into actionable tasks for specialized agents. How we built our multi-agent research system dives even deeper into how to build with these orchestrators and the design elements required to make them effective. Essentially Anthropic describes an "augmented LLM"—a language model enhanced with external tools and resources. It also acts as the main interface between the user and the system in the context of MCP it is referred to as the client.
Rather than building our orchestrator from scratch, we'll leverage a powerful existing coding agent Claude Code (Alternatives are GitHub Copilot or Continue as an open source option). There are many options to choose from and we can even spin up our own ReAct agent in DSPy. Choosing a coding agent is a strategic choice - these agents are already built to do things like file manipulation and code writing. By using a powerful commercial model as the central 'reasoning engine,' we can focus our efforts on building highly specialized pharmacometric tools.
Note that in the diagram above, the agent architecture includes some of the very well designed tools built into Claude Codes, such as file management, todo list management, and the ability to parallelize and launch subagents to complete tasks. Not all coding agents have these features, but in that case we could connect an MCP server to the orchestrator to provide similar functionality.
Building Our First Tool: The Analysis Plan Parser with DSPy
To demonstrate this architecture, we'll build our first tool: an LLM that parses an analysis plan using DSPy. DSPy represents a paradigm shift from traditional prompting to programming with language models. We define our LLM logic as signatures and modules which can be composed, reused, and optimized (more on optimization in the next post).
Here's how we build our extract_tasks_tool:
import dspy
from langchain_community.document_loaders import Docx2txtLoader
from pydantic import BaseModel, Field
# Define the structure of the task and task list using Pydantic models
class Task(BaseModel):
"""Structure for pharmacometric analysis tasks"""
task: str = Field(description="Short name for the analysis")
description: str = Field(description="Step by step breakdown of how the task will be performed")
class ExtractTasksSignature(dspy.Signature):
"""Extract analysis tasks from an analysis plan."""
text: str = dspy.InputField(
description="Text content of the analysis plan document, typically extracted from a .docx file"
)
task_list: list[dict[str, str]] = dspy.OutputField(
description="List of dictionaries of tasks extracted from the analysis plan. Each list item contains a dictionary with keys 'task' and 'description' with the corresponding string values.",
)
module = dspy.Predict(ExtractTasksSignature)
def extract_tasks(filename: str) -> str:
"""Extract analysis tasks from an analysis plan."""
# Load the document using Docx2txtLoader
loader = Docx2txtLoader(filename)
text = loader.load()
# Extract the text content from the loaded document
response = module(text=text)
# Unpack the task list to a markdown string, using the first and second keys dynamically
task_list = response.task_list
task_list_md = "\n".join(
[
f"### {list(task.values())[0]}\n{list(task.values())[1]}" if len(task) >= 2 else str(task)
for task in task_list
]
)
return task_list_md
if __name__ == "__main__":
# Initialize DSPy
lm = dspy.LM('ollama_chat/qwen2.5-coder:7b', api_base='http://localhost:11434', api_key='', temperature=0.9)
dspy.configure(lm=lm)
# Example usage
filename = "./data/analysis_plan_arimab.docx"
tasks = extract_tasks(filename)
print("Extracted Tasks:")
print(tasks)This module produces a structured, machine-readable JSON object, providing reliable output to be used as input for downstream code generation and analysis agents. We've built a tool i.e., a python function. We could bind this directly to our orchestrator agent as a tool or wrap this as an MCP server. Wrapping it allows universal integration with any MCP-compliant agent and has the advantage of running the tool server-side. This means we don't have to burden the orchestrator with the logic of how to run the tool, just the input/output.
We can deploy this tool as an MCP server, allowing our orchestrator to call it with a simple script as follows from the FastMCP documentation:
import dspy
from fastmcp import FastMCP
from src.tools import TOOLS
# Initialize the server
mcp = FastMCP(name="PharmAI", tools=TOOLS)
# Initialize DSPy
lm = dspy.LM('ollama_chat/qwen2.5-coder:7b', api_base='http://localhost:11434', api_key='', temperature=0.9)
dspy.configure(lm=lm)
if __name__ == "__main__":
# Start the server
mcp.run()We then install this into our client (Claude Code) with a json config file (.mcp.json):
{
"mcpServers": {
"PharmAI": {
"command": "/path/to/uv",
"args": [
"--directory",
"/path/to/PharmAI/mcp_server.py",
"run",
"python",
"mcp_server.py"
]
}
}
}Once set up, our interaction with Claude Code looks like this:
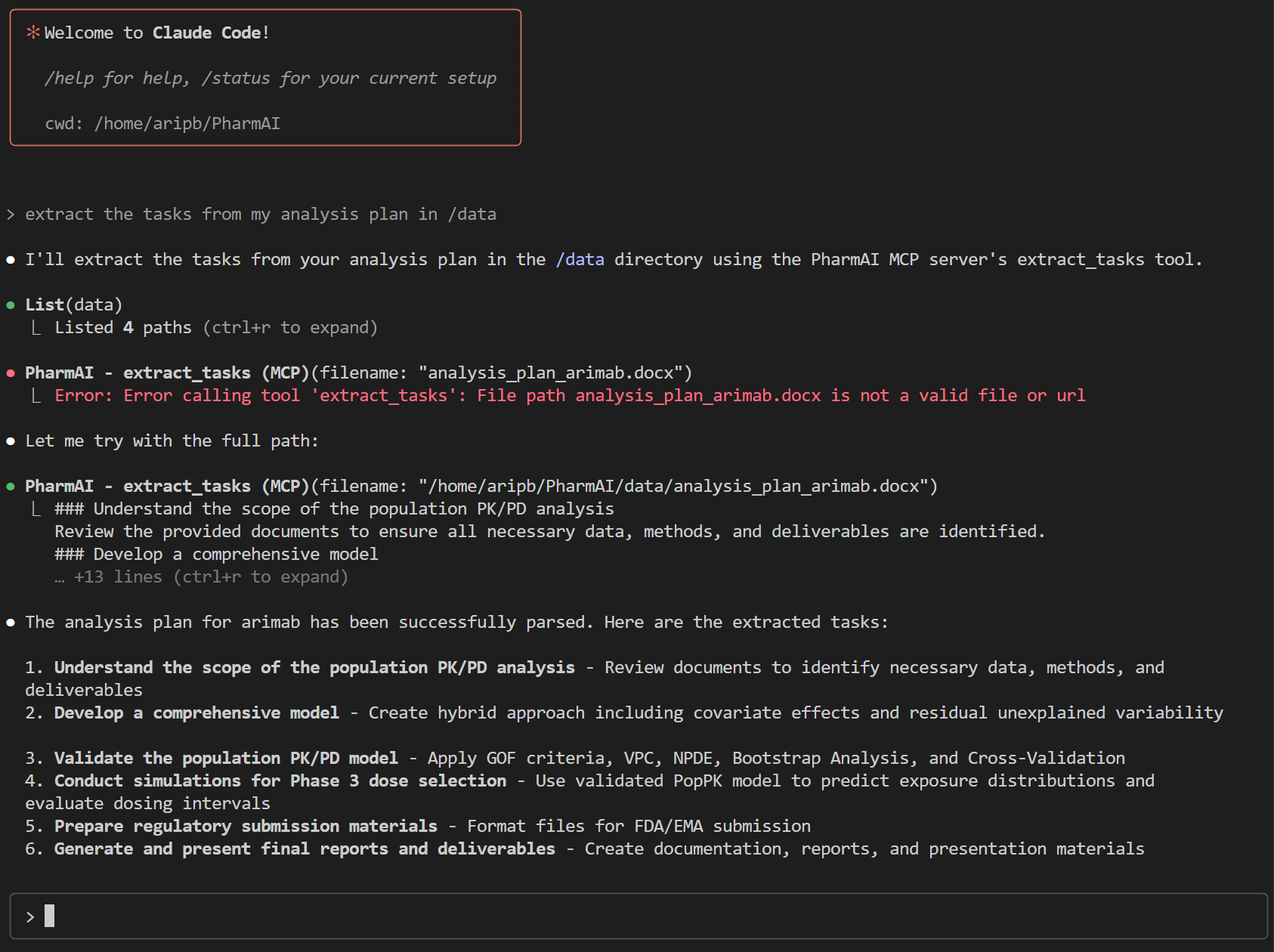
Claude Code will query our MCP server, which would run the extract_tasks_tool, returning a structured JSON object with the tasks extracted from the document. I liked this example in particular because it shows a simple error recovery by Claude Code (the orchestrator) when it didn't pass the correct file location. It then corrected itself and successfully called the MCP server to extract the tasks.
This is a modular step that could either be chained into a more complex task by the orchestrator or even another MCP server that handles task execution in a secure environment such as Code Sandbox MCP.
Thanks to IndyDevDan for great tutorials on how to set up MCP servers with Claude Code and FastMCP.
Tying it All Together: The Full System Architecture
The following figure illustrates how these components interact in our simplified system:
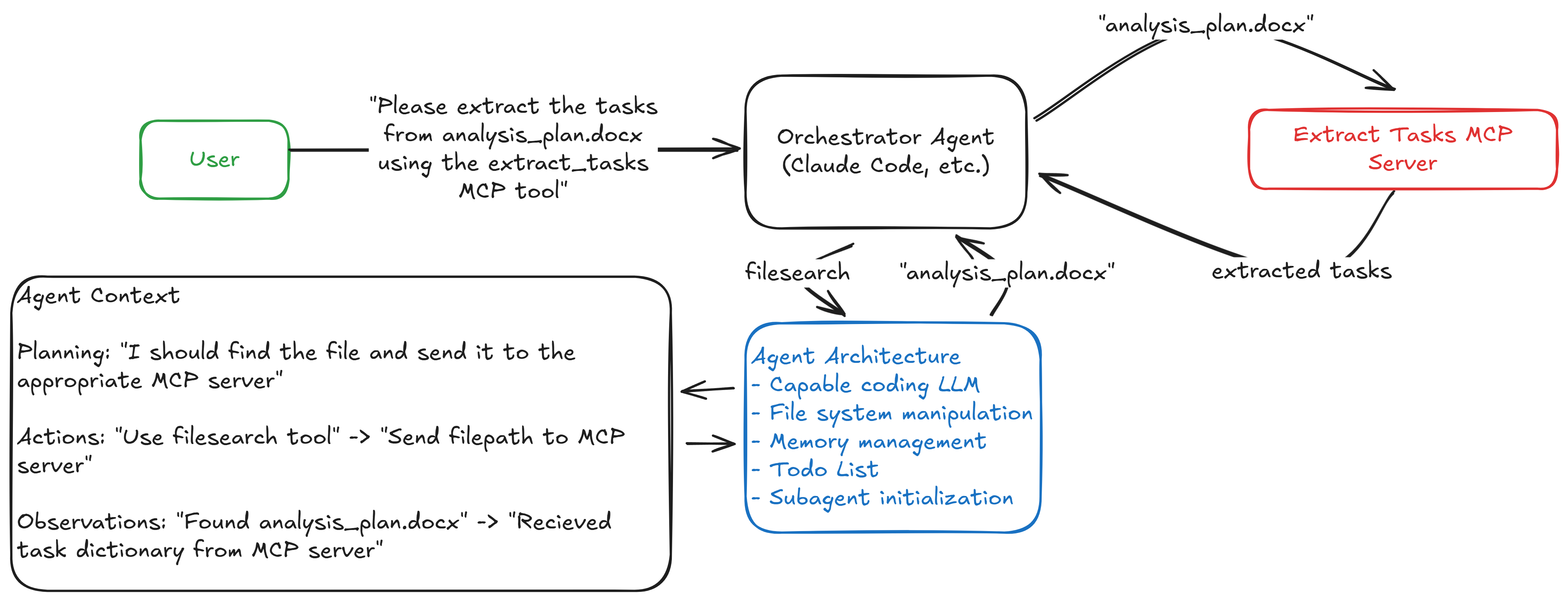
The orchestrator receives a high-level goal and uses simple text commands to systematically call specialized MCP or internal tools. Each tool performs its function while the orchestrator maintains the overall workflow and context.
Wrapping Up: What We Accomplished and Where We're Going
Let's recap what we built today. We designed the blueprint for a scalable, multi-agent pharmacometric system, moving from a single monolithic agent to a composable architecture of specialized tools. This is critical for building the robust, maintainable, and validatable systems required for regulated environments.
Key Takeaways from This Post:
- Composability over Monoliths: A system of specialized, independent tools managed by an orchestrator is more scalable, robust, and easier to validate than a single, all-powerful agent.
- MCP as the Communication Backbone: The Model Context Protocol (MCP) provides a standard, plug-and-play interface for connecting our orchestrator to various specialized tools and agents.
- DSPy for Building Robust Tools: We can use DSPy to "program" our language models, creating reliable, structured outputs (like our
extract_tasks_tool) which can be optimized.
The work we've done here provides the perfect foundation for our next challenge. Now that we have a framework for building specialized tools, we are ready to think about optimizing them.
In the next post, we will focus on building robust evaluation frameworks ("evals") for our pharmacometric tools. We'll explore how to decompose full pharmacometric workflows into subtasks and create high-quality datasets for systematically improving our tools' performance via prompt optimization and reinforcement learning.
You can find a fully implemented extract_tasks_tool, along with the MCP server integration code, in our PharmAI GitHub repository. Share your thoughts in the comments or reach out on LinkedIn. Better yet, join me in this community effort to build open-source pharmacometric AI tools. Together, we can transform how we work with complex data and models in this field.
This post was developed with assistance from AI. All code examples are provided as educational material and should be thoroughly tested before production use. Views expressed are my own and do not represent my employer.