Why This Post Exists
Next week, I'll be presenting at the American Conference on Pharmacometrics (ACoP 2025) in two formats: a poster session on Sunday, October 19th, and a brief talk on Monday, October 20th as part of a quality award ceremony. This post serves as a detailed companion to those presentations, a reference document for anyone who wants to dive deeper into the architecture, principles, and code behind AI-driven pharmacometric workflows.
The abstract and poster can be found on the ACoP website. (Note: you'll need to be signed in to view the poster.)
If you're attending ACoP, I'd love to connect. If you're not, this post will give you the full picture of what we're building and why it matters.
The Journey So Far
For the past two years, I've been building an AI automation system for pharmacometric analysis at a large pharmaceutical company. That work has led to an ongoing blog series exploring different aspects of this challenge:
- Part 1: The Vision: Why AI in pharmacometrics requires secure, auditable, human-in-the-loop systems
- Part 2: The Foundation: Building a single-agent framework for R script generation
- Part 3: The Orchestration: Multi-agent architecture with DSPy and Model Context Protocol
- Part 4: The REPL: Self-correcting loops for iterative debugging
This post represents an evolution of that work and introduces the architecture I'm presenting at ACoP. It's simpler, more robust, and more production-ready than what I've shown before.
Who This Is For
This post is written for pharmacometricians and AI engineers working in or curious about pharmaceutical applications. If you've used ChatGPT for basic tasks but wonder how to build reliable, auditable systems for complex scientific workflows, this is for you.
I share the code for this project in our PharmAI GitHub repository (recent updates pending).
The Architecture: An Overview
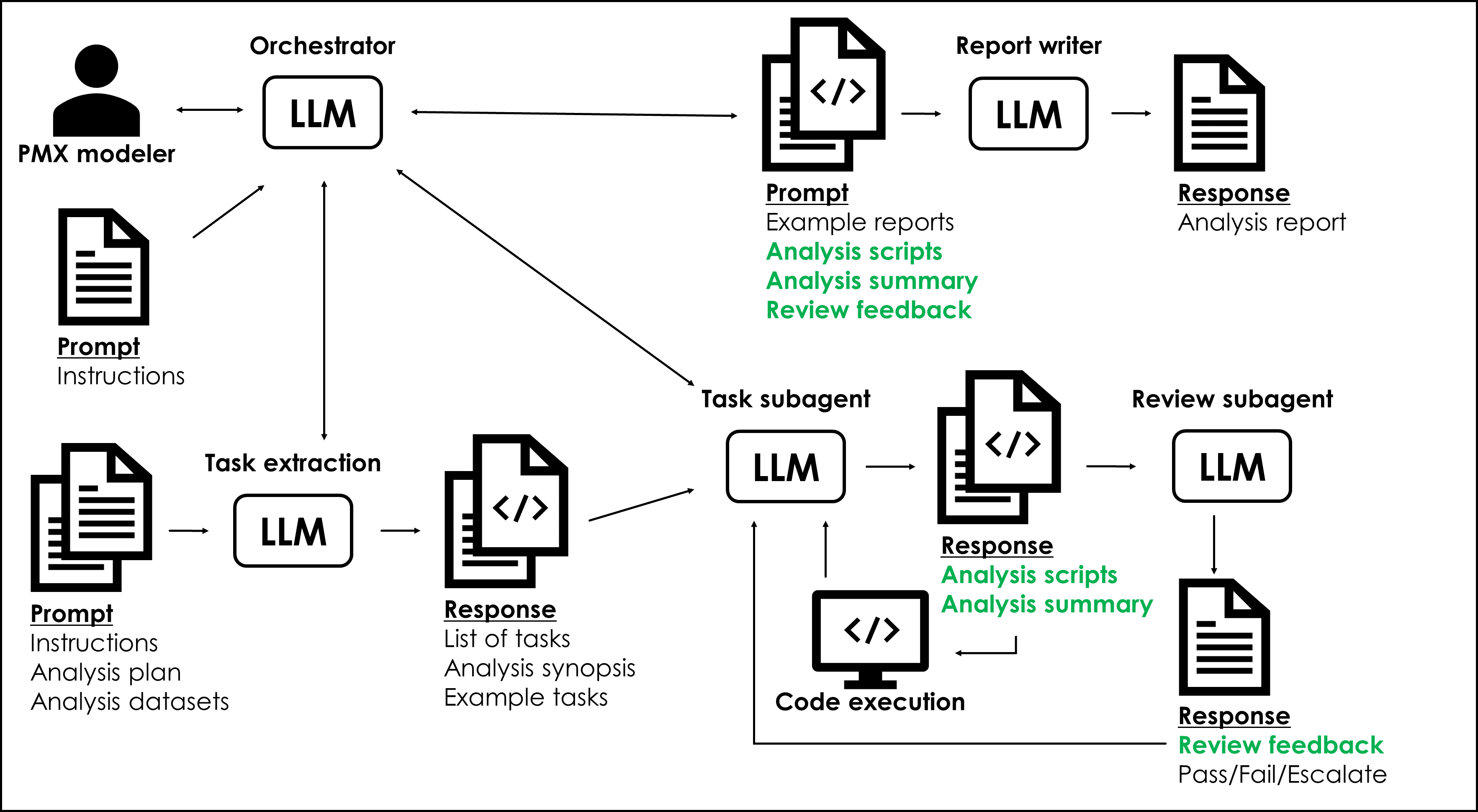
The system uses Claude Code as the foundation, with a main orchestrator agent that delegates work to specialized subagents. Each subagent operates in its own isolated context window and is optimized for a specific type of pharmacometric task.
The workflow has six key components:
- Orchestrator - The main Claude Code instance that coordinates everything
- Task Parser - Extracts structured tasks from analysis plans
- Specialized Subagents - Domain-specific agents for EDA, structural modeling, and mixed-effects modeling
- Reviewer Subagent - Quality control and validation
- Reporter Subagent - Aggregates results and generates structured reports
Understanding Claude Code Subagents
Before diving into the specifics, it's worth understanding what subagents are and why they matter for this type of system.
What Are Subagents?
In Claude Code, subagents are specialized AI assistants defined as Markdown files with YAML frontmatter. They live in your project's .claude/agents/ directory. Each one has:
- A name and description that helps the orchestrator understand when to delegate to them
- Custom system prompts defining their expertise, behavior, and constraints
- Tool access that can be restricted to only what they need (Read, Write, Bash, etc.)
- Isolated context windows so they maintain separate memory from the main conversation
Example Subagent Structure:
---
name: eda-agent
description: Performs exploratory data analysis and dataset preparation for pharmacometric modeling. Use when you need data cleaning, visualization, or preliminary analysis.
tools: Read, Write, Bash, Grep
---
You are an expert in pharmacometric data analysis with deep knowledge of CDISC standards, PK/PD data structures, and exploratory visualization techniques.
Your role is to generate R code for data cleaning, exploratory analysis, and visualization of pharmacokinetic and pharmacodynamic data.
Read the example scripts in the examples/ directory to understand expected code patterns and outputs.
Expected outputs:
1. Cleaned dataset in NONMEM format
2. Exploratory plots (concentration-time, distribution summaries)
3. Data quality report highlighting any issues
Use dplyr and tidyr for data manipulation, ggplot2 for visualization, and PKNCA for non-compartmental analysis.Why Subagents Matter for Pharmacometrics
Subagents provide two critical advantages.
First, context isolation. Each subagent works in its own context window. When your EDA agent reads a 50,000-row dataset and generates diagnostic plots, all that detailed output stays in its context. The orchestrator only receives the summary and key findings, keeping the main workflow focused and efficient.
Second, domain specialization. By giving each subagent example scripts, domain-specific knowledge, and clear constraints, they become experts in their specific task. The structural modeling agent doesn't need to know about data cleaning, and the EDA agent doesn't need to understand mixed-effects estimation algorithms.
1. The Orchestrator: Coordination Through Custom Commands
The orchestrator is a Claude Code instance configured through a custom slash command. Slash commands in Claude Code are Markdown files in .claude/commands/ that define specific behaviors and workflows.
When you launch Claude Code with the orchestrator command (like /pmx-orchestrator), it receives the analysis plan from the user, delegates to the task parser to extract structured tasks, spawns specialized subagents based on task types, monitors progress and coordinates handoffs between subagents, and interfaces with the user for approvals and feedback.
The orchestrator maintains the high-level workflow but doesn't do the detailed work itself. That's what the subagents are for.
Example Orchestrator Command Structure (.claude/commands/pmx-orchestrator.md):
---
description: Orchestrates multi-agent pharmacometric analysis workflow
---
You are the orchestrator for a pharmacometric analysis system. Your role is to coordinate specialized subagents to complete complex analyses.
Workflow:
1. Parse the analysis plan using the task extractor CLI tool
2. For each task, determine which subagent to invoke:
- EDA/dataset tasks → eda-agent
- Structural modeling → structural-pkpd-agent
- Mixed-effects modeling → mixed-effects-agent
3. After each task, invoke the reviewer-agent for QC
4. Once all tasks complete, invoke the reporter-agent for final synthesis
Your responsibilities:
- Maintain context of overall analysis goals
- Ensure task dependencies are respected
- Communicate with user for approvals
- Track progress and handle errors
Do not write code yourself. Delegate to specialized subagents.How It Works in Practice
When the orchestrator runs, it sets up a structured workspace with a clear directory hierarchy. Each task gets its own subdirectory containing the code, outputs, and review results. The main directory includes a task list JSON file that tracks progress and dependencies. This file system organization ensures everything is traceable and auditable, which is critical for regulated work. You can see at a glance what each subagent produced, what passed review, and what needed iteration.
2. Task Parsing: From Natural Language to Structured Work
The entry point for any analysis is the analysis plan, a document describing objectives, datasets, and modeling approaches. The task parser extracts executable tasks from this plan.
The task extractor is a standalone Python CLI script that uses a DSPy module for structured parsing. The orchestrator calls this script directly as a command-line tool. I kept DSPy for task extraction because it benefits from having structured, optimizable signatures. This is the bridge between natural language and the structured workflow the subagents need.
Example Output (Structured Tasks JSON):
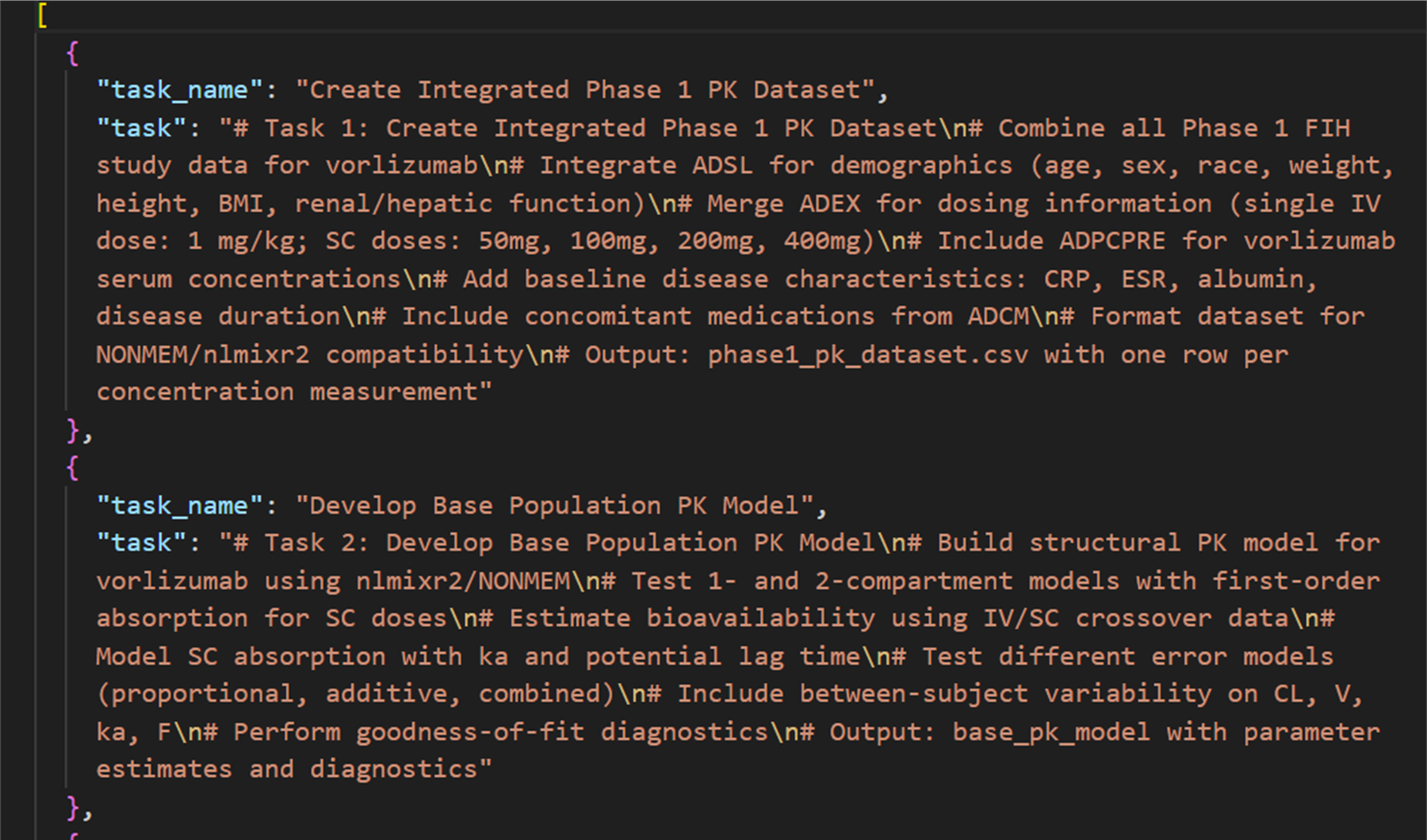
Once tasks are extracted, the orchestrator then decides which subagents to invoke and in what order.
3. Specialized Subagents: The Workers
Each specialized subagent is pre-configured with domain-specific system prompts explaining its role, a dedicated folder containing example R scripts, and clear input/output specifications. When a subagent is invoked, it's instructed to read the example scripts in its folder to understand expected patterns and best practices.
For example, the EDA subagent has an examples/eda/ directory with working scripts showing data cleaning workflows, standard diagnostic plots, and output formatting. The subagent reads these as reference material before generating code for its specific task.
The Three Core Subagents
Below are the general roles and example configurations for each of the three main subagents. Note that in production these would be more detailed and refined. Eventually these are tuned and optimized through evals and feedback loops.
PharmEDA Subagent
Handles data preparation, cleaning, and exploratory visualization.
Subagent Configuration (.claude/agents/pharmEDA.md):
---
name: pharmEDA
description: Performs exploratory data analysis and dataset preparation. Use for data cleaning, visualization, and preliminary PK/PD analysis.
tools: Read, Write, Bash
---
You are an expert in pharmacometric data analysis.
Read the example scripts in examples/eda/ to understand standard patterns for data cleaning and visualization.
Your tasks:
- Receive raw dataset specification and task description
- Generate R code using dplyr, ggplot2, and PKNCA
- Produce diagnostic plots (concentration-time curves, distribution summaries)
- Output a cleaned dataset ready for modelingExample Task Execution:
Orchestrator: "Use the eda-agent to generate concentration-time plots stratified by dose"
[eda-agent spawns with its own context]
[Reads example scripts from examples/eda/]
[Reads the data file]
[Generates R script]
[Executes code and produces plots]
[Returns summary: "Completed EDA. 3 plots generated, dataset cleaned, 247 subjects included"]
Orchestrator receives only the summary, not the 50KB of plot data and logs.PharmStructural: PKPD Modeling Subagent
Performs individual and/or naive pooled fitting with grid search to identify reasonable starting parameter values.
This subagent uses grid search to explore parameter space, fits models using base R optimization (optim, nls), evaluates goodness-of-fit, and outputs parameter estimates for the mixed-effects stage.
Before jumping into population modeling, we need physiologically reasonable starting values. The structural model provides this foundation. Analytical equations are shown below, however, we've also given the agent example scripts using the fantastic package mrgsolve to simulate and fit one-compartment models with first-order absorption.
Example Generated Code Snippet:
# One-compartment model with first-order absorption
pk_model <- function(params, time, dose) {
ka <- params[1]
ke <- params[2]
V <- params[3]
conc <- (dose / V) * (ka / (ka - ke)) *
(exp(-ke * time) - exp(-ka * time))
return(conc)
}
# Grid search over parameter space
param_grid <- expand.grid(
ka = seq(0.1, 2, by = 0.1),
ke = seq(0.01, 0.5, by = 0.05),
V = seq(10, 100, by = 10)
)
# Fit and evaluate across grid
best_params <- find_best_fit(param_grid, observed_data)PharmModeler: Mixed Effects Modeling Subagent
Builds population models using another amazing open-source package nlmixr2, incorporating inter-individual variability and covariates.
This subagent translates the structural model into nlmixr2 syntax, adds random effects on key parameters, tests covariate relationships, and generates VPC plots, GOF diagnostics, and parameter estimates.
Example Generated Code Snippet:
library(nlmixr2)
pk_popmod <- function() {
ini({
tka <- 0.5; label("Absorption rate constant")
tcl <- 4; label("Clearance")
tv <- 70; label("Volume of distribution")
eta.ka ~ 0.1
eta.cl ~ 0.1
eta.v ~ 0.1
add.sd <- 0.1
})
model({
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
ke <- cl / v
d/dt(depot) <- -ka * depot
d/dt(central) <- ka * depot - ke * central
cp <- central / v
cp ~ add(add.sd)
})
}
fit <- nlmixr(pk_popmod, data = pk_data, est = "saem")4. The REPL Loop: Built-In Iterative Debugging
One of Claude Code's most powerful features is that iterative debugging happens automatically. Each subagent operates in a Read-Eval-Print-Loop (REPL) by default:
- Read: Parse the task and context
- Eval: Generate and execute R code
- Print: Check results, errors, and diagnostics
- Loop: If errors occur, debug and retry
This isn't something we had to engineer ourselves. The incredible team at Anthropic has built Claude Code to maximize agentic capabilities, which includes running and iterating on code to accomplish a task. When a subagent encounters an error (wrong column name, missing package, unexpected data structure), it reads the error message, understands what went wrong, modifies the code, re-executes, and continues until success or reaches a limit.
This self-correction is critical for production reliability. Real-world datasets are messy, and hardcoded assumptions break. The REPL loop handles this gracefully.
5. The Reviewer Subagent: Automated Quality Control
After each task completes, the orchestrator delegates to the reviewer subagent for quality assurance.
It checks whether the code ran without errors, whether outputs are reasonable (parameter estimates within physiologically plausible ranges), whether diagnostic plots are present and interpretable, and whether the output meets the task requirements.
The output is a structured "go/no-go" decision with detailed feedback.
Example Review:
{
"task_id": 3,
"status": "PASS",
"feedback": "Population PK model successfully fit. Parameter estimates are physiologically reasonable (CL = 4.2 L/hr, V = 68 L). VPC shows good model fit with observations within 90% prediction intervals. Covariate effects on clearance are statistically significant.",
"warnings": ["Residual error slightly heteroscedastic - consider proportional error model in next iteration"]
}The reviewer also has an escalation mechanism. If it determines that the analysis has gone completely off track or encounters issues beyond its ability to resolve, it can escalate back to the orchestrator, which then prompts the human modeler for input. This human-in-the-loop checkpoint ensures that the system doesn't continue down an incorrect path without intervention.
The reviewer operates in its own context, so it can deeply analyze outputs without cluttering the orchestrator's workflow.
6. The Reporter Subagent: Synthesis and Communication
The final step is synthesis. The reporter subagent aggregates task summaries from each specialized subagent, review feedback from the reviewer, and key outputs (plots, tables, parameter estimates).
It generates a structured report that a human pharmacometrician can review, approve, or request modifications. Because the report is grounded in actual analysis scripts that were executed, with direct references to the code and outputs, there's minimal risk of hallucination. The reporter can only describe what actually happened in the analysis.
Key Principles: What We Learned Building This
1. The Bitter Lesson: Less Structure is Better
Richard Sutton's "The Bitter Lesson" from AI research states that methods relying on general computation (search, learning) tend to outperform methods relying on human-designed structure.
What this meant in practice: don't over-engineer the orchestration layer, let Claude Code handle delegation naturally. Provide examples, not rigid rules. Give subagents example scripts and let them generalize. Trust the REPL loop because iterative debugging works better than trying to generate perfect code upfront.
This matters especially now because the technology is evolving rapidly. The length of tasks these agents can handle is roughly doubling every seven months. Right now, language models can complete an hour-long task with 50% success rate. In seven months, that doubles. We're building on top of technology that's fundamentally improving underneath us, so minimizing rigid structure lets us benefit from these improvements automatically.
Moving from custom MCP/DSPy orchestration to Claude Code's native patterns was a simplification that improved reliability.
2. Context Engineering: Specific I/O Specifications
Each subagent is initialized with clear input specifications (what data format, what task description), expected output format (what files, what structure, what metrics), and example reference scripts (working R code demonstrating best practices stored in dedicated folders).
This isn't traditional prompt engineering. It's context engineering. We're setting up the environment so each agent has everything it needs to succeed without polluting the main workflow's context.
3. Evals: Quantitative Evaluation at Every Step
To build trust, we need quantitative evaluation at each stage. Here are concrete examples:
- Task Extraction: Compare the extracted tasks against what we expect from the analysis plan. Did it capture all the required analyses?
- Dataset Building: Check if all subjects from the original dataset are accounted for. Are the data transformations correct?
- Model Diagnostics: Are GOF plots within acceptable ranges? Are parameter estimates physiologically plausible?
- Decision Making: Run retrospective studies where we know the outcome. Does the system make the same clinical decisions as expert modelers, or even better ones?
We use these retrospective studies (where we know the "right answer") to test and refine our evaluation criteria. This is how we move from proof-of-concept to production.
Why Pharmacometrics is Uniquely Suited for AI Automation
Two factors make pharmacometrics an ideal domain for AI-driven workflows.
1. Rich Software Ecosystem
Pharmacometrics has mature, well-documented tools: nlmixr2 for population modeling, mrgsolve for clinical trial simulation, PKNCA for non-compartmental analysis, and ggplot2 for visualization.
These tools have clear interfaces, extensive documentation, and established best practices. AI agents can learn to use them effectively because there's a wealth of example code and tutorials.
What's particularly powerful is that agents can learn any new software tool on the fly. When new open-source packages are released (and many are being presented right here at ACoP), they can be integrated directly into the agent workflow by simply providing documentation and examples. The system isn't hardcoded to specific tools. It adapts to the ecosystem as it evolves.
Note to ACoP attendees: The developers of these packages are in the audience. Thank you! This system is built on your work.
2. Infinite Synthetic Data Generation
Unlike many domains, pharmacometrics can generate infinite synthetic data using model-based clinical trial simulation. We can test the system exhaustively on known-answer scenarios. We can refine and optimize evals without privacy concerns. We can create adversarial examples (edge cases, corrupted data) to stress-test reliability.
This is a massive advantage. We're not limited by small datasets or regulatory constraints during development.
What This Means for the Field
This architecture represents a shift from custom-built orchestration to leveraging production-ready tools for the hard parts. Claude Code's native subagent system, REPL loops, and context isolation solve problems we were previously engineering around.
The takeaways: Claude Code orchestrator uses custom slash commands to coordinate the workflow. Specialized subagents are defined as Markdown files, each with domain expertise, example script folders, and isolated context. Automatic REPL debugging means iterative error correction is built in. Reviewer and reporter subagents provide automated QC and structured reporting for audit trails. The pharmacometrics-specific advantages of a rich software ecosystem plus synthetic data generation make this domain particularly well-suited for AI automation.
The result is simpler, more maintainable, and more reliable than earlier custom implementations.
Let's Connect at ACoP
If you're attending ACoP, come find me at the poster session on Sunday, October 19th, or at my talk on Monday, October 20th (8-minute presentation during the quality award ceremony).
I'll give the full architecture diagram walkthrough, and have plenty of time to discuss implementation details, challenges, and your specific use cases.
If you're not at ACoP but want to discuss AI automation in pharmacometrics, reach out. I'm building this in the open, and I'd love to hear your challenges, ideas, and skepticism.
What's the biggest barrier you see to adopting AI in your pharmacometric workflows? Reach out on LinkedIn or check out the PharmAI GitHub repo.
This post was developed with assistance from AI. Views expressed are my own and do not represent my employer.